Noam Shental sent me the following:
Together with my colleagues, Amnon Amir (Physics of Complex System, Weizmann Institute Of Science, Israel) and Or Zuk (Broad Institute, Boston), we are currently working on a Compressed Sensing (CS) approach to a problem in genomics, namely the detection of rare genetic mutations in large populations.As far as we know, our work is the first to apply CS in the context of next-generation sequencing technology in genomics. As we are not CS experts, we would highly appreciate comments from people in this field. A draft of our article has been submitted to the arxiv (http://arxiv.org/abs/0909.0400), and any feedback will be very welcome. We also believe that this application may be of interest to people in the CS community.
The paper is: Rare-Allele Detection Using Compressed Se(que)nsing by Noam Shental, Amnon Amir and Or Zuk. The abstract reads:
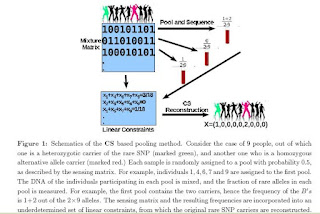
Detection of rare variants by resequencing is important for the identification of individuals carrying disease variants. Rapid sequencing by new technologies enables low-cost resequencing of target regions, although it is still prohibitive to test more than a few individuals. In order to improve cost trade-offs, it has recently been suggested to apply pooling designs which enable the detection of carriers of rare alleles in groups of individuals. However, this was shown to hold only for a relatively low number of individuals in a pool, and requires the design of pooling schemes for particular cases. We propose a novel pooling design, based on a compressed sensing approach, which is both general, simple and efficient. We model the experimental procedure and show via computer simulations that it enables the recovery of rare allele carriers out of larger groups than were possible before, especially in situations where high coverage is obtained for each individual. Our approach can also be combined with barcoding techniques to enhance performance and provide a feasible solution based on current resequencing costs. For example, when targeting a small enough genomic region (∼100 base-pairs) and using only ∼ 10 sequencing lanes and ∼10 distinct barcodes, one can recover the identity of 4 rare allele carriers out of a population of over 4000 individuals.
This work looks very similar to the work done by Raghu Kainkaryam and Anna Gilbert using sparse random matrices to perform high throughput testing as mentioned here, here and here. Raghu then mentioned to me that:
I don't know if you saw Nicolas Thierry-Mieg's recent paper on applying the STD pooling strategy (the equivalent of DeVore's deterministic constructions of CS matrices) to protein-protein interaction mapping:http://genome.cshlp.org/content/19/7/1262.abstract
After further investigation, Nicolas then mentioned to me that this paper is available here. It is: Shifted Transversal Design smart-pooling for high coverage interactome mapping by Xiaofeng Xin, Jean-Francois Rual, Tomoko Hirozane-Kishikawa, David E. Hill, Marc Vidal, Charles Boone, and Nicolas Thierry-Mieg. The abstract reads:
‘‘Smart-pooling,’’ in which test reagents are multiplexed in a highly redundant manner, is a promising strategy for achieving high efficiency, sensitivity, and specificity in systems-level projects. However, previous applications relied on low redundancy designs that do not leverage the full potential of smart-pooling, and more powerful theoretical constructions, such as the Shifted Transversal Design (STD), lack experimental validation. Here we evaluate STD smartpooling in yeast two-hybrid (Y2H) interactome mapping. We employed two STD designs and two established methods to perform ORFeome-wide Y2H screens with 12 baits. We found that STD pooling achieves similar levels of sensitivity and specificity as one-on-one array-based Y2H, while the costs and workloads are divided by three. The screening-sequencing approach is the most cost- and labor-efficient, yet STD identifies about twofold more interactions. Screening-sequencing remains an appropriate method for quickly producing low coverage interactomes, while STD pooling appears as the method of choice for obtaining maps with higher coverage.
Unrelated to these papers, an example of sparsity in everything I mentioned yesterday are the non-functioning Protein-Protein Interactions.
No comments:
Post a Comment